Early
farming village in China (Wikicommons – Xinyang City Museum, Gary Todd)
Respiratory
viruses began to propagate more easily when hunting and gathering gave way to
farming and as settlements grew larger. Humans may have then evolved to use coronaviruses
as a natural vaccine against deadlier respiratory diseases, like tuberculosis
and pneumonia.
A
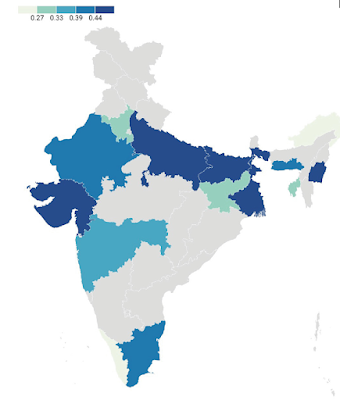
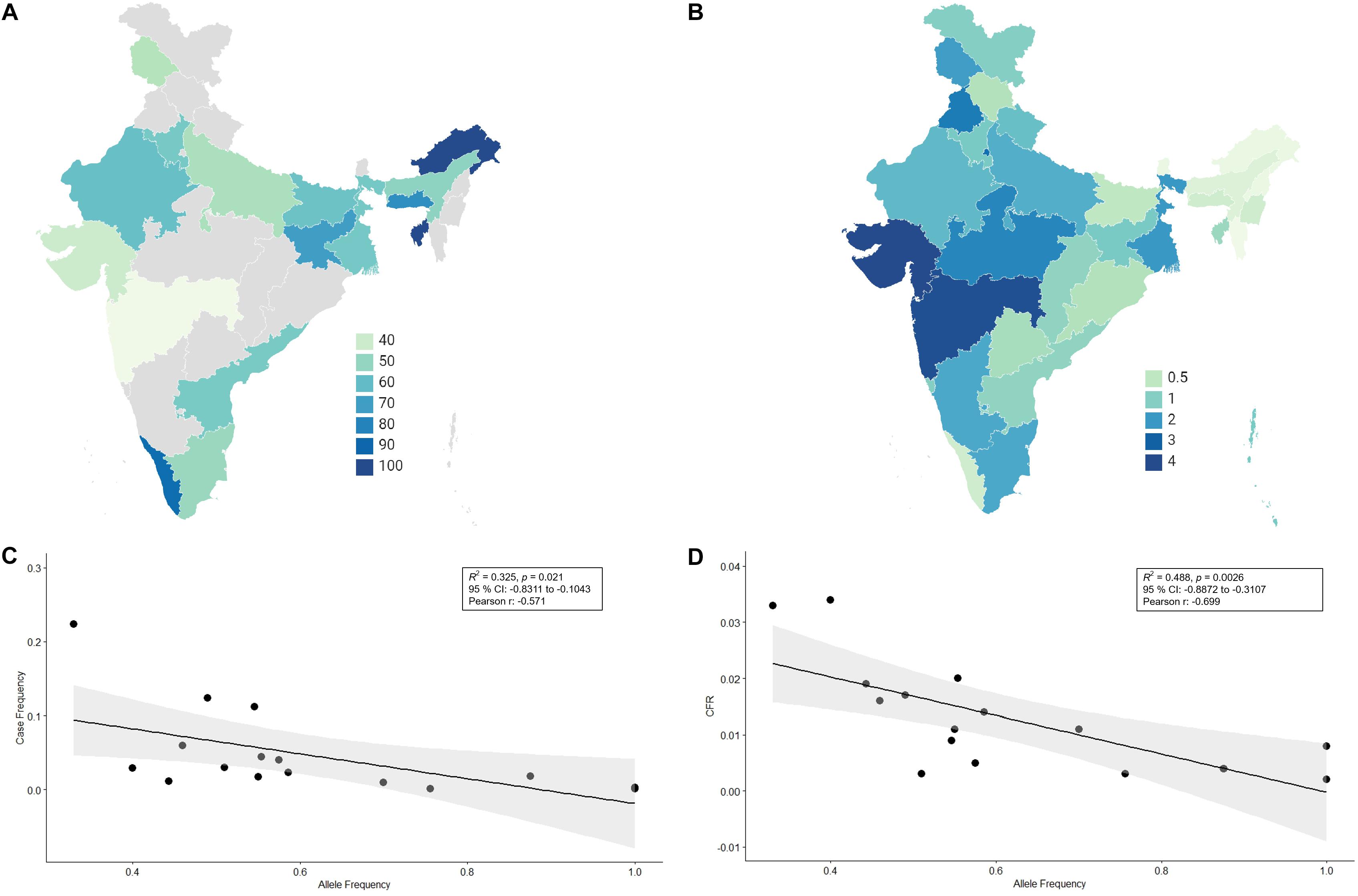
new genomic study has found that East Asians had to adapt to epidemics of coronaviruses
some 25,000 years ago. The authors looked at gene variants for proteins that
interact with coronaviruses in five East Asian populations: Han Chinese
(Beijing); Han Chinese (South China); Dai (Yunnan, China); Japanese; and
Vietnamese. There were three main findings:
·
Ancestral
East Asians had to adapt to coronavirus epidemics around 25,000 years ago
·
They
adapted by acquiring mutations that are close to genes that regulate the
development of lung tissue and other tissues affected by COVID-19
·
Those
mutations either promote or block infection by coronaviruses (Souilmi et al.
2021, p. 3505).
The
last finding is puzzling. Did those ancestral East Asians become more vulnerable
or less vulnerable to coronaviruses? The authors simply say that half of those
mutations from 25,000 years ago have “anti- or proviral effects” versus 29% of
all proteins that interact with coronaviruses (Souilmi et al. 2021, p. 3509).
Fine. But how many of those mutations were antiviral and how many proviral?
It
might seem strange that natural selection would actually make people more susceptible
to coronavirus infections. Yet such susceptibility could be beneficial. A viral
infection can boost immunity to other pathogens, including deadly ones that
cause tuberculosis, pneumonia, or pneumonic plague. Until recently, coronaviruses
were typically mild in their effects, producing what we call the “common cold.”
They may thus act as a natural vaccine against deadlier respiratory diseases
(Frost 2020).
Respiratory diseases are believed to have become serious for humans when hunting and
gathering gave way to farming. People became sedentary and their settlements grew
larger with time, thus providing respiratory viruses with better conditions for
propagation (Comas et al. 2013). This theoretical model is in conflict,
however, with the above finding that ancestral East Asians began adapting to
coronaviruses some 25,000 years ago, long before they adopted farming and became
sedentary. We’re thus left with the unlikely conclusion that coronavirus
epidemics began among scattered bands of hunter-gatherers.
The
estimate of 25,000 years ago is probably wrong. The authors arrived at that figure
by calculating the latest date when the ancestors of the four East Asian groups
were still a single population. But East Asians are not descended from a single
population. Their origins are best described by the "Two-Layer" (TL)
hypothesis:
·
Modern
humans spread into East Asia through a northern route and a southern route.
·
The
southerners were then replaced to varying degrees by northerners who spread out
of northeast Asia and successively occupied northern China, southern China, and
Southeast Asia (Oxenham and Buckley 2016; Xu et al. 2006).
·
Thus,
as you go farther south in East Asia, the population has a greater admixture
from the earlier southern “layer”—from hunter-gatherers who closely resemble
the relic groups that still exist in parts of Southeast Asia, i.e., the Andaman
Islanders, the Aeta of the Philippines and the Maniq and Semang of the Malayan
Peninsula.
Admixture
from that older southern substrate pushes back in time the latest common ancestors,
who never existed. Adaptation to
coronaviruses therefore happened at a later date, probably when the
“northerners” pushed into what is now northern China and adopted farming. They
then grew in population, pushed farther south, and intermixed with the
hunter-gatherers who lived there.
References
Comas,
I., M. Coscolla, T. Luo, et al. (2013). Out-of-Africa migration and
Neolithic coexpansion of Mycobacterium tuberculosis with modern
humans. Nature Genetics 45:
1176–1182. https://doi.org/10.1038/ng.2744
Frost,
P. (2020). Does a commensal relationship exist between coronaviruses and some
human populations? Journal of
Molecular Genetics 3(2): 1-2. https://researchopenworld.com/does-a-commensal-relationship-exist-between-coronaviruses-and-some-human-populations/
Frost,
P. (2022). A natural vaccine. Evo and
Proud, February 21 http://evoandproud.blogspot.com/2022/02/a-natural-vaccine.html
Oxenham,
M., and H.R. Buckley. (2016). The population history of mainland and island
Southeast Asia, in M. Oxenham and H.R. Buckley (eds) The Routledge Handbook of Bioarchaeology in Southeast Asia and the
Pacific Islands. Routledge.
Souilmi,
Y., M.E. Lauterbur, R. Tobler, C.D. Huber, A.S. Johar, S.V. Moradi, W.A.
Johnston, N.J. Krogan, K. Alexandrov, and D. Enard. (2021). An ancient viral
epidemic involving host coronavirus interacting genes more than 20,000 years
ago in East Asia. Current Biology
31(16), 3504–3514.e9. https://doi.org/10.1016/j.cub.2021.05.067
Xue,
Y., T. Zerjal, W. Bao, S. Zhu, Q. Shu, J. Xu, R. Du, S. Fu., P. Li, M.E.
Hurles, H. Yang, C. Tyler-Smith. (2006). Male demography in East Asia: A
north-south contrast in human population expansion times. Genetics 172: 2431-2439, https://doi.org/10.1534/genetics.105.054270